Abstract
Acute myeloid leukemia (AML) makes up nearly 20% of acute pediatric leukemias and has a long-term survival rate of approximately 70%. A key driver of the low overall survival for children with AML is relapsed disease and less than 40% of the children with relapsed AML will survive past five years. We recently characterized relapsed pediatric AML (pAML) to better understand the spectrum of genomic alterations at disease recurrence (Umeda et al., 2022). However, the genome-wide patterns of genetic evolution from diagnosis to relapse have yet to be characterized for pAML. Thus, there is a gap in knowledge of how alterations at diagnosis evolve during treatment to drive relapse and poor outcome.
We performed whole genome sequencing (WGS) and whole transcriptome sequencing (RNA-Seq) on a cohort of 50 pediatric AML patients with paired diagnosis and relapse samples. The median age at diagnosis was 10.7 yrs (range 0.7-20yrs), and the median time between diagnosis and relapse was 334 days (range 93-1,773 days). The main driver group contained a KMT2A fusion event (n=16, 32%). NUP98 fusion made up the second largest group at 14% (n=7). Also within the cohort are newly defined categories, including a BCL11B structural variant (n=1) and patients with UBTF tandem duplication AML (n=4). These features are overall similar to the molecular profiles of pediatric AML (Farrar et al., 2016; Umeda et al., 2022).
Approximately 22,500 alterations (including point mutations, indels, and structural variants) were detected by WGS (median coverage 36X). The median number of alterations at diagnosis and relapse per patient was, respectively, 224 (range 118-867) and 411 (range 73-1,909). Most of the cohort noticed more alterations at relapse than diagnosis, but some (n=8) had fewer at relapse. Additionally, there was no trend with the time to relapse and the number of acquired alterations. The most commonly mutated genes regardless of timepoint included FLT3 (n=16, 6 were internal tandem duplications), WT1 (n=18), NRAS (n=16), and SETD2 (n=9). We identified over 11,000 relapse enriched variants (median 169, range 3-1,378), including alterations that had a significantly higher allele frequency (q-value < 0.05) at relapse compared to diagnosis. This included 1,028 relapse enriched variants that alter protein coding genes. FLT3 contained 7, 2, and 7 relapse enriched, relapse depleted, and time point stable alterations, respectively. WT1 contained 6, 2, and 10 relapse enriched, relapse depleted, and time point stable alterations, respectively. Furthermore, the relapse-enriched list included three SETD2 protein-truncating mutations and four AUTS2 structural variants often co-occurring with KMT2A fusions, indicating that these mutations may cooperate with KMT2A fusion and contribute to the disease progression.
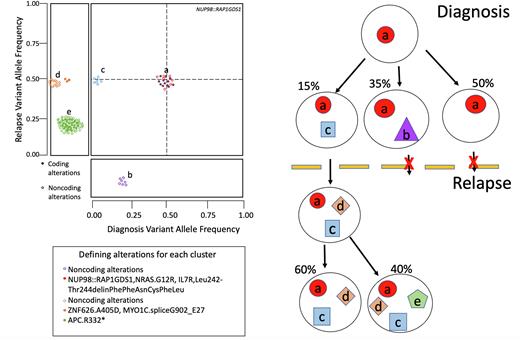
To increase our understanding of the clonal structure, 36 cases with additional DNA material were re-sequenced with a custom panel covering 6,300 variants, including all coding variants for each case. This led to a validation rate of 97% of all variants and the increased sequencing depth of the panel (median 265X for diagnosis and 274X for relapse) gave greater insight into the changes in clonal structure between diagnosis and relapse time points. Most of the cohort (70%) featured a relapsed enriched clone that was minor at diagnosis and became clonal in relapse. Additional cases (n=10) featured a clone that was undetectable by WGS at diagnosis and after treatment became the dominant clone in relapse. Only one case maintained the same major clone in both diagnosis and relapse.
In summary, our data show a comprehensive characterization of changes in the clonal structure of pAML between diagnosis and relapse, revealing relapse-enriched gene alterations which may be driving relapse disease.
Farrar, J. E., Schuback, H. L., Ries, R. E., Wai, D., Hampton, O. A., Trevino, L. R., . . . Meshinchi, S. (2016). Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res, 76(8), 2197-2205. doi:10.1158/0008-5472.CAN-15-1015
Umeda, M., Ma, J., Huang, B. J., Hagiwara, K., Westover, T., Abdelhamed, S., . . . Klco, J. M. (2022). Integrated genomic analysis identifies UBTF tandem duplications as a recurrent lesion in pediatric acute myeloid leukemia. Blood Cancer Discov. doi:10.1158/2643-3230.BCD-21-0160
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal